Table of Contents
Definition / general | Essential features | Terminology | ICD coding | Epidemiology | Sites | Pathophysiology | Etiology | Clinical features | Diagnosis | Laboratory | Prognostic factors | Case reports | Treatment | Microscopic (histologic) description | Microscopic (histologic) images | Peripheral smear description | Peripheral smear images | Molecular / cytogenetics description | Molecular / cytogenetics images | Sample pathology report | Differential diagnosis | Additional references | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Younes IE, Zhang L. PDGFRB rearrangement. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/myeloproliferativePDGFRB.html. Accessed April 19th, 2024.
Definition / general
- Myeloid or lymphoid neoplasm with frequent eosinophilia, sometimes neutrophilia or monocytosis; associated with rearrangement of PDGFRB, at chromosome 5, long arm (5q32)
- Discovered in 1994 by Golub TR, Barker GF, Lovett M, Gilliland DG (Cell 1994;77:307)
- To date, more than 40 fusion partners identified
Essential features
- Can have different presentation; e.g., myeloproliferative neoplasm (MPN), myelodysplastic syndrome / myeloproliferative neoplasm (MDS / MPN), acute myeloid leukemia (AML), with MDS / MPN overlap syndrome being more common
- Conventional karyotyping is the most common used modality for diagnosis, confirmation by FISH PDGFRB (break apart probe) or RT-PCR is recommended due to multiple other genes being located at 5q31-33 (e.g., IL3, IL5, GM-CSF)
Terminology
- Chronic myelomonocytic leukemia with eosinophilia associated with t(5;12); myeloid neoplasms with PDGFRB rearrangement
- Myeloid neoplasms associated with PDGFRB rearrangement
ICD coding
- ICD-11: 2A51 - myeloid neoplasm associated with PDGFRB rearrangement
Epidemiology
- M:F = 2:1
- Age range is 8 - 72 years with peak incidence at late 40s (Acta Haematol 2002;107:113)
Sites
- Peripheral blood and bone marrow is always involved
- Spleen; other organs can be involved as well
Pathophysiology
- Most common translocation t(5;12)(q32;p13.2) ETV6::PDGFRB fusion gene
- More than 40 different partners are identified (Histopathology 2020;76:1042)
- t(1;3;5)(p36;p21;q33) WDR48::PDGFRB
- t(1;5)(q21;q33) TPM3::PDGFRB
- t(1;5)(q23;q33) PDE4DIP::PDGFRB
- t(5;7)(q33;q11.2) HIP1::PDGFRB
- t(5;10)(q33;q21) CCDC6 (H4/D10S170)::PDGFRB
- Some fusions are typically associated with BCR::ABL1-like B cell acute lymphoblastic leukemia (B ALL) (e.g., PDGFRB fused with EBF1, SSBP2, TNIP1, ZEB2 and ATF7IP)
- Those fusions are not diagnosed as PDGFRB rearranged entities but rather as BCR::ABL1-like B ALL (N Engl J Med 2014;371:1005)
Etiology
- Unknown
Clinical features
- Can present with MPN, MDS / MPN (e.g., atypical chronic myeloid leukemia [CML], chronic myelomonocytic leukemia [CMML] or juvenile myelomonocytic leukemia [JMML])
- Rarely myeloid blast phase / secondary AML or de novo B ALL (Histopathology 2020;76:1042, Blood 2017;129:704)
- Can present with B symptoms
- Hepatosplenomegaly
- Cardiac and skin damage
- Good response to imatinib or other tyrosine kinase inhibitor (TKI) therapy
Diagnosis
- Peripheral blood and bone marrow examination both usually show eosinophilia (sometimes normal) and other features representative of MPN, MDS / MPN or transformed AML
- Conventional karyotyping
- FISH PDGFRB for confirmation, especially cryptic cases
- RT-PCR has limited utility due to multiple fusion partners (Foucar: Bone Marrow Pathology, 4th Edition, 2019)
- Next generation sequencing (NGS) can detect cases missed by FISH (Cancer Genet 2020;244:55)
- According to WHO, diagnostic criteria for myeloid / lymphoid neoplasms associated with ETV6::PDGFRB or other rearrangement of PDGFRB:
- Myeloid or lymphoid neoplasm often with prominent eosinophilia and sometimes with neutrophilia or monocytosis and presence of t(5;12)(q32;p13.2) or a variant translocationa,b or demonstration of ETV6::PDGFRB fusion gene or other rearrangement of PDGFRBb
- Cases with fusion genes typically associated only with BCR::ABL1-like B lymphoblastic leukemia are specifically excluded
- Because t(5;12)(q32;p13.2) does not always result in ETV6::PDGFRB fusion, molecular confirmation is highly desirable; if molecular analysis is not possible, this diagnosis should be suspected if there is a myeloproliferative neoplasm associated with eosinophilia, with no Philadelphia (Ph) chromosome and with a translocation with a 5q32 breakpoint
- Myeloid or lymphoid neoplasm often with prominent eosinophilia and sometimes with neutrophilia or monocytosis and presence of t(5;12)(q32;p13.2) or a variant translocationa,b or demonstration of ETV6::PDGFRB fusion gene or other rearrangement of PDGFRBb
Laboratory
- Complete blood count (CBC) usually shows eosinophilia; however, normal counts can be found
- Increased tryptase levels (> 12 ng/dL); however, < 20 ng/dL (Expert Rev Hematol 2014;7:683)
- Vitamin B12 levels may be increased
Prognostic factors
- Median overall survival is < 2 years before introduction of tyrosine kinase inhibitor; a case series of 26 cases treated with imatinib showed a longer survival (up to 6.6 years)
Case reports
- 4 month old girl with refractory juvenile xanthogranuloma with a novel MRC1::PDGFRB fusion and a dramatic response induced by dasatinib (Blood Adv 2020;4:2991)
- 22 year old woman with relapsed BCR::ABL1-like ALL with CCDC88C::PDGFRB fusion and complete remission after treatment with tyrosine kinase inhibitor (Int J Hematol 2021;113:285)
- 22 year old man with a cryptic imatinib sensitive G3BP1::PDGFRB rearrangement in a myeloid neoplasm with eosinophilia (Blood Adv 2020;4:445)
- 70 year old man with asymptomatic leukocytosis, thrombocytosis with PDGFRB rearrangement and absence of eosinophilia (Leukemia 2016;30:1402)
- 76 year old man with myeloid / lymphoid neoplasms associated with eosinophilia, with negative FISH and positive next generation sequencing for PDGFRB rearrangement (Cancer Genet 2020;244:55)
Treatment
- Sensitive to tyrosine kinase inhibitor (TKI) (e.g., imatinib)
- Study showed a response rate to imatinib treatment of 96% with a 10 year overall survival rate of 90% (Blood 2014;123:3574, Blood 2017;129:704)
- Some mutations confer resistance to all TKIs (e.g., PDGFRB C843G and EBF1::PDGFRB Ph-like ALL with gatekeeper mutation T681I) (Blood 2018;131:2256, Haematologica 2021;106:2242)
Microscopic (histologic) description
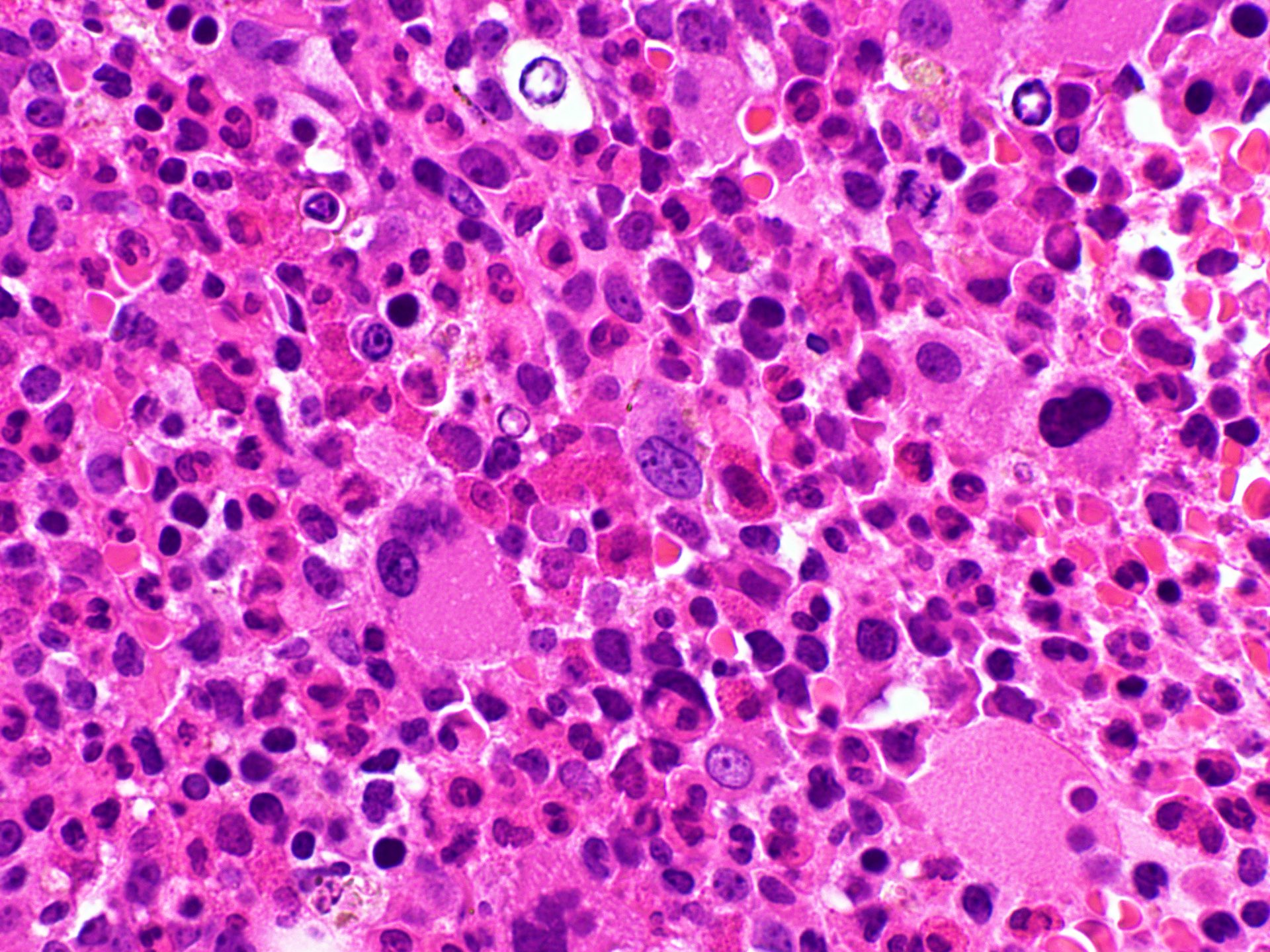
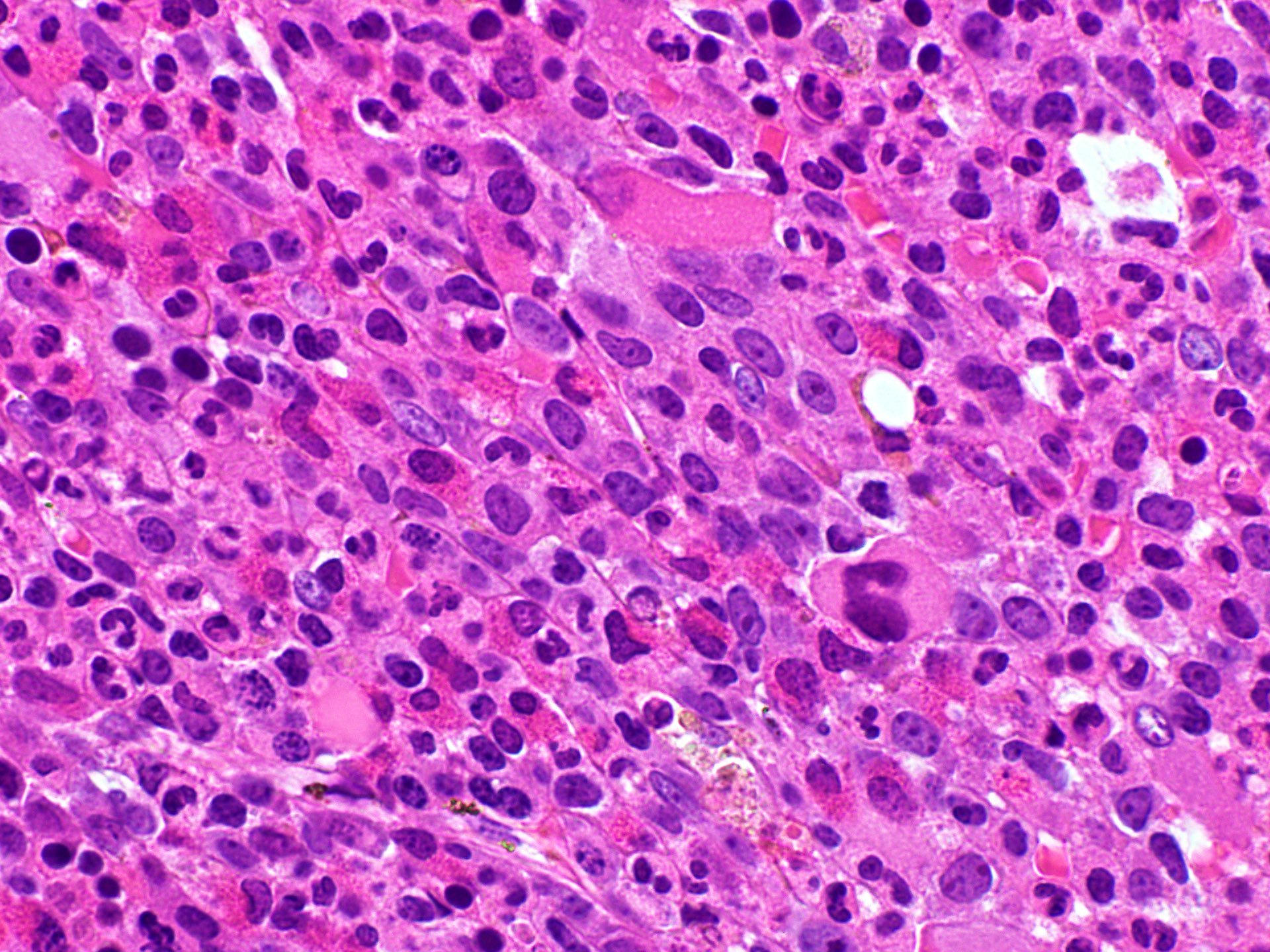
- Peripheral blood shows leukocytosis including eosinophilia, neutrophilia or monocytosis; sometimes circulating immature myeloid precursors (e.g., metamyelocytes, myelocytes or promyelocytes and occasional blasts / blast equivalents)
- Increased blasts up to 20% seen in blastic phage
- Anemia and thrombocytopenia can be seen
- Bone marrow is hypercellular with myeloid preponderance including many eosinophils and neutrophils
- Rarely there might be increase in basophils
- Can show increase in mast cells with identified abnormal spindle shaped forms and expression of CD2 and CD25 (Am J Hematol 2007;82:77, Haematologica 2007;92:163)
- Reticulin fibrosis (Blood 2008;111:1855)
Microscopic (histologic) images
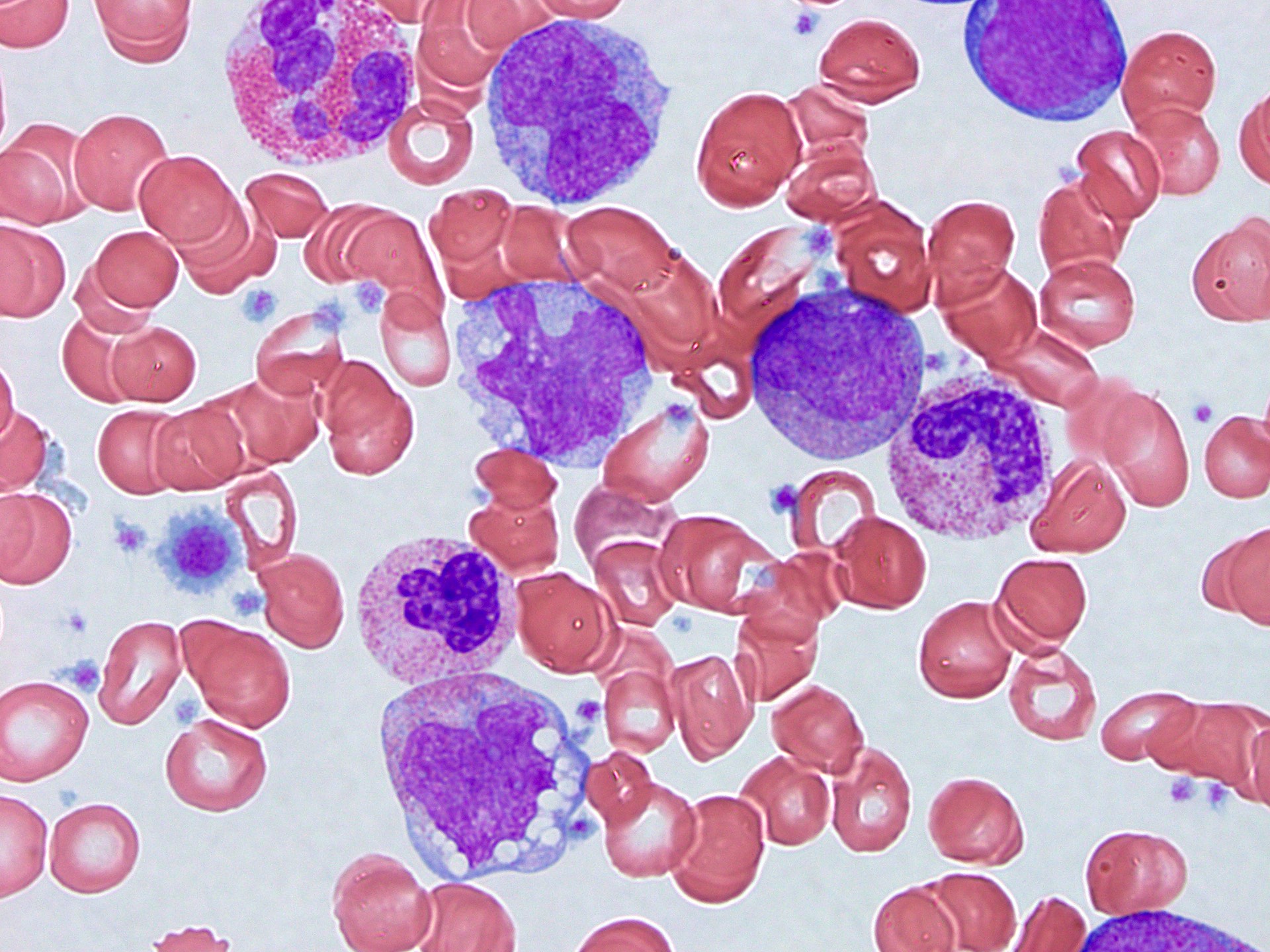
Peripheral smear description
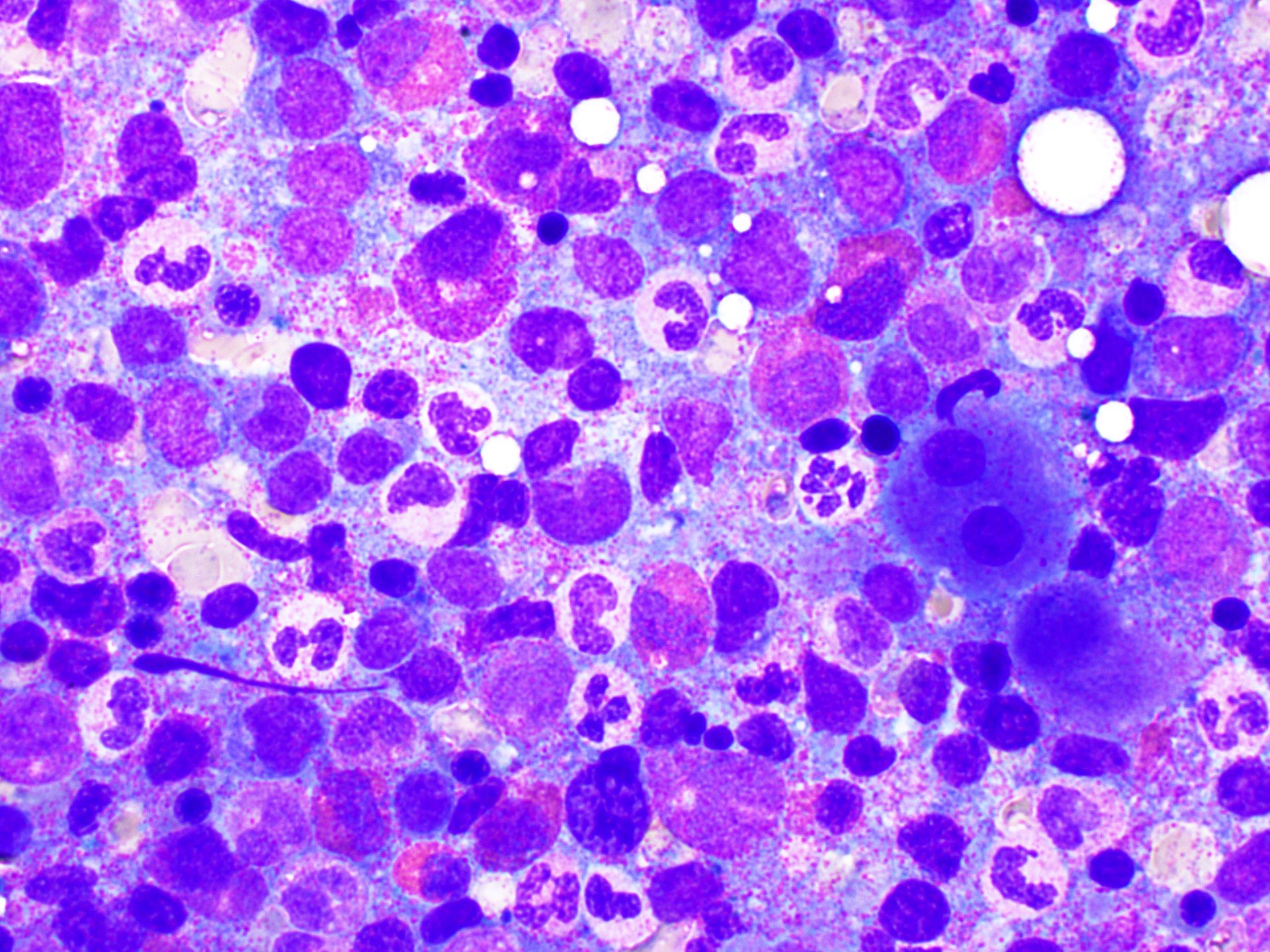
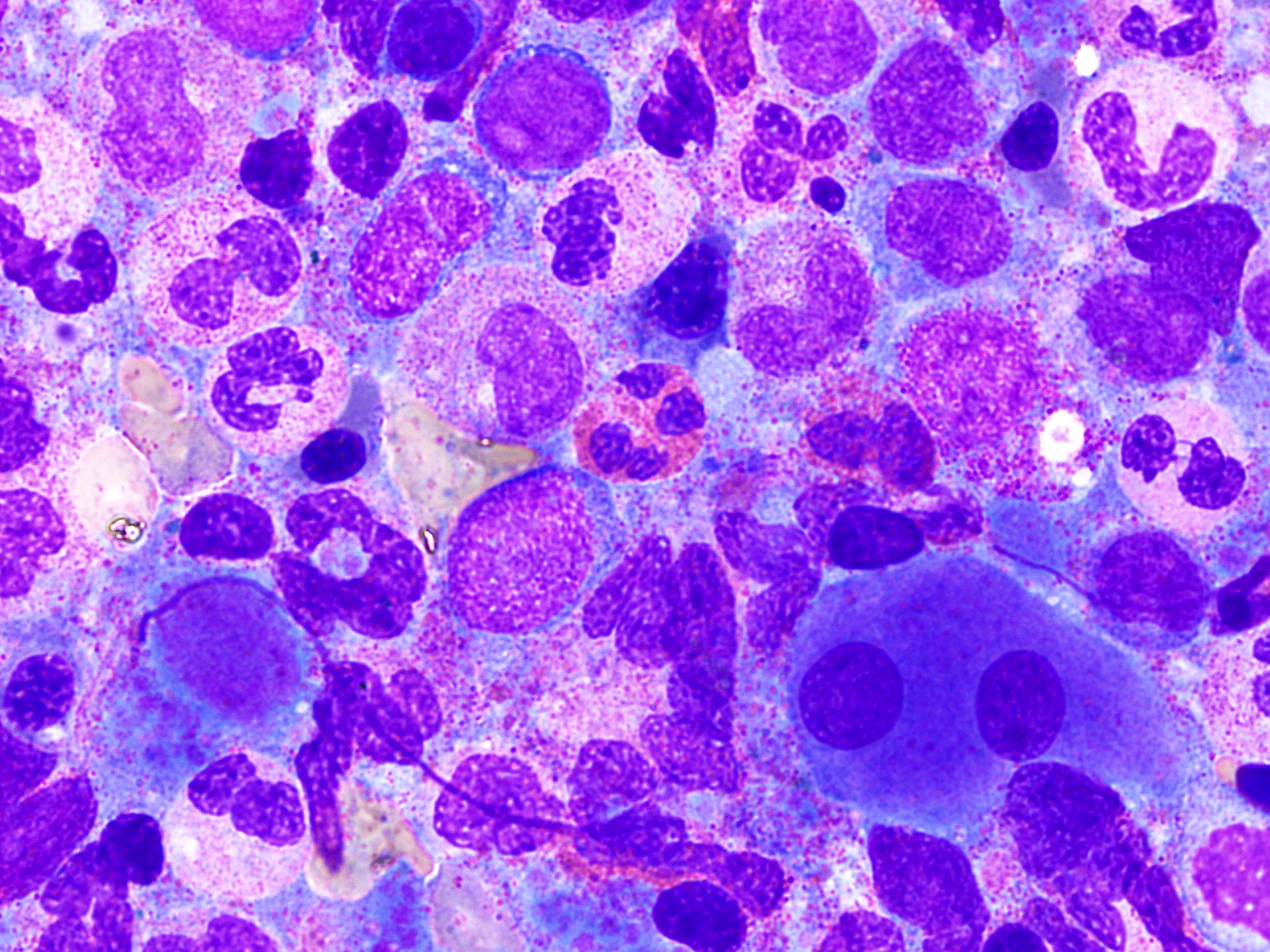
- Peripheral blood eosinophilia and occasionally neutrophilia, eosinophils are usually mature
- Variable morphologic abnormalities can be present in the eosinophils, including cytoplasmic vacuolation, nuclear hyper / hyposegmentation
- Morphologic changes are not unique as reactive conditions can have these changes as well (Foucar: Bone Marrow Pathology, 4th Edition, 2019)
Peripheral smear images
Molecular / cytogenetics description
- FISH rearrangement with 5q31~33 breakpoint, typically t(5;12)(q31~33;p12) creating ETV6::PDGFRB
Molecular / cytogenetics images
Sample pathology report
- Peripheral blood:
- Mild macrocytic anemia
- Neutrophilia with left shifted granulocyte maturation
- Monocytosis
- Mild thrombocytopenia
- Bone marrow, left posterior iliac crest, aspirate and biopsy:
- Hypercellular marrow with myeloid hyperplasia and monocytosis, consistent with a myeloid neoplasm with PDGFRB rearrangement (see comment)
- Mildly increased reticulin fibers
- Comment: Cytogenetics reported 46,XY,t(5;14)(q33;q32)[18]/46,XY[2]. FISH was reported positive for PDGFRB gene rearrangement. An MDS FISH panel and FISH for BCR::ABL were reported negative. Flow cytometry reported atypical myeloid maturation with decreased CD10, atypical maturation of CD64 / CD14 and increased HLA-DR expression. Given the cytogenetic findings, this myeloid neoplasm with PDGFRB rearrangement is associated with a hematologic diagnosis of chronic myelomonocytic leukemia with eosinophilia. The morphology is compatible with chronic myelomonocytic leukemia and there are focal areas of slightly increased eosinophils seen on the core biopsy.
- Microscopic description:
- Peripheral smear: There is a mild macrocytic anemia with mild anisopoikilocytosis. The total white blood cell count is elevated, with a neutrophilia and monocytosis. Left shifted granulocytes are seen, including an occasional circulating blast. The monocytes are maturing. The granulocytes have variable cytoplasmic granulation. Platelets are mildly decreased.
- Aspirate smear: The Wright stained aspirate smear is cellular, with spicules available for evaluation. Megakaryocytes are seen in adequate number, with an occasional hypolobated form. Myeloid and erythroid precursors are seen. The myeloid precursors show slight left shifted maturation, with approximately 4% blasts. The myeloid precursors have variable toxic granulation and include a few precursors with mild nuclear:cytoplasmic asynchrony. Monocytes comprise approximately 25% of the differential and are unevenly distributed. The monocytes include approximately 5% promonocytes and many immature monocytes. Eosinophils include forms with some large basophilic granules. Erythroid precursors show maturation. Lymphocytes and scattered plasma cells are seen in the background.
- Aspirate iron: An aspirate smear is stained for iron by the contributor. Stainable iron stores are present. A rare ring sideroblast is seen.
- Clot section and core biopsy: Subcortical bone and normal bony trabeculae surround the marrow space with a cellularity approaching 100%. Megakaryocytes are seen in slightly decreased numbers, with occasional hypolobated forms. Small erythroid islands with admixed myeloid precursors are seen, with an M:E ratio estimated at greater than 5:1. The myeloid precursors have focal areas of left shifted maturation. No clusters of blasts are seen on the H&E stained section. Focal areas of slightly increased eosinophils are noted. Stainable iron stores are adequate. Reticulin fibers are mildly increased.
Differential diagnosis
- Chronic eosinophilic leukemia, NOS:
- Sustained eosinophilia (> 1.5 x 109/L) with other reactive processes excluded
- < 20% blasts in peripheral blood or bone marrow, no inv(16)(p13.1q22), t(16;16)(p13.1q22), t(8;21)(q22;q22.1)
- Bone marrow aspirate is hypercellular with abundant eosinophils and eosinophilic precursors with dysplastic features
- Clonal cytogenetic (e.g., +8, -20q) or molecular genetic abnormalities (e.g., TET2, ASXL1 and DNMT3A)
- If no clonal abnormalities are found, then blast count must be > 2% in peripheral blood or > 5% in bone marrow aspirate
- No PDGFRA, PDGFRB, FGFR rearrangement, BCR::ABL1, PCM1::JAK2, ETV6::JAK2 or BCR::JAK2 fusions
- No lymphoid or myeloid neoplasms (e.g., MDS, MPNs, systemic mastocytosis [SM] or AML) are identified
- Hypereosinophilic syndrome:
- Sustained eosinophilia (> 1.5 x 109/L) for at least 6 months with other reactive processes excluded
- Tissue damage must be present
- Exclude cytokine related lymphoid variant hypereosinophilia
- Bone marrow shows increased eosinophils with no dysplastic features and no increase in blast cells
- No cytogenetic or molecular alteration is identified
- No PDGFRA, PDGFRB, FGFR rearrangement, BCR::ABL1, PCM1::JAK2, ETV6::JAK2 or BCR::JAK2 fusions
- No inv(16)(p13.1q22), t(16;16)(p13.1q22) or t(8;21)(q22;q22.1)
- No lymphoid or myeloid neoplasms (e.g., MDS, MPNs, SM or AML) are identified
- Reactive eosinophilia:
- Can present with increased eosinophils and can have abnormal eosinophils in the peripheral blood
- Secondary causes are present: infection (e.g., parasites; the most common cause), connective tissue disorders, certain cutaneous lesions (e.g., dermatitis herpetiform), paraneoplastic (e.g., Hodgkin lymphoma, T cell lymphoma, colon carcinoma), certain lung diseases (e.g., Churg-Strauss syndrome, aspergillosis), autoimmune disorder or on IL2 therapy (Hematology Am Soc Hematol Educ Program 2018;2018:326)
- No evidence of PDGFRB rearrangement
- Idiopathic hypereosinophilia:
- Sustained eosinophilia (> 1.5 x 109/L) for at least 6 months
- No tissue damage
- No reactive causes, lymphoid or myeloid neoplasms are identified
- Bone marrow shows increased eosinophils with no dysplastic features and no increase in blast cells
- No molecular alteration is identified
- No PDGFRA, PDGFRB, FGFR rearrangement, BCR::ABL1, PCM1::JAK2, ETV6::JAK2 or BCR::JAK2 fusions
- Systemic mastocytosis with eosinophilia:
- Major diagnostic criteria: 4 minor criteria or multifocal infiltrates of mast cells (≥ 15 mast cells in aggregates) + 1 minor criterion
- Minor criteria
- Bone marrow shows eosinophilia with variable dysplastic features
- No PDGFRB rearrangement
Additional references
Board review style question #1
Board review style answer #1
Board review style question #2
Which of the following is the least useful in diagnosing in myeloid / lymphoid neoplasms with eosinophilia and PDGFRB rearrangement?
- FISH
- Karyotyping
- RT-PCR
- Snap array
Board review style answer #2