Table of Contents
Definition / general | Essential features | Terminology | Developing a virtual staining algorithm | Evaluation of virtual stains | Diagrams / tables | Registration software | Virtual staining software | Select example applications of virtual stains | Advantages and disadvantages | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Levy J, Vaickus L. Virtual staining. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/informaticsvirtualstaining.html. Accessed April 18th, 2024.
Definition / general
- Virtual staining (VS) is the conversion of an H&E (or other common standard stain, e.g., PAS) digital whole slide image (WSI) to a digital prediction of a special stain (e.g., trichrome, CK AE1 / AE3 IHC)
- The prediction relies on the detection of morphological features (if they exist) of the source WSI, which inform the distribution of a special stain
- Typically performed using artificial neural networks (ANN)
- Can obviate the need for application of physical reagents, thereby conserving precious laboratory resources (e.g., reagents, staff time, costs) while improving operational efficiency (e.g., view multiple stains simultaneously via nonautonomous diagnostic decision aid) and standardization of reagents (i.e., assessment quality)
- It can also serve as a quick initial assessment while waiting for a physical special stain to be processed
Essential features
- Virtual staining technologies leverage ANNs to generate synthetic images of tissue slides with predictions of the results of special staining reagents (chemical and IHC)
- Bolsters pathology laboratory infrastructure through rapid reflexive staining at virtually no cost
- Selection of stains is application dependent and whether the suspected histomorphology provides sufficient information to infer marker
- 2 primary approaches exist for generation of synthetic stain:
- Use of a generative adversarial network to produce image of synthesized stain
- Identify and segment cells which have been tagged by coregistered IHC / mIF WSI on same section
- Validated visually using Turing test (ability to distinguish real and synthetic), qualitative ratings and clinical concordance to diagnostic / prognostic outcomes
Terminology
- Whole slide image (WSI): digitized representation of tissue slide after whole slide scanning at 20x or 40x resolution (e.g., using Aperio AT2 or GT450 scanner); slide dimensionality can exceed 100,000 pixels in any given spatial dimension
- Subimage / patch: smaller, local rectangular region extracted from a WSI, often done to ameliorate use of excessive computational resources that may hinder development and deployment of machine learning algorithm (J Pathol Inform 2019;10:9)
- Immunohistochemistry (IHC): application of chemical staining reagents with selective antibodies that bind to specific antigens to assess for the presence of particular biomarkers
- Multiplexed immunofluoresence (mIF): multispectral imaging after excitation of multiple fluorophores attached to antibodies (Cancer Commun (Lond) 2020;40:135)
- Special stains: alternative staining techniques which are not explicitly routine (e.g., H&E) or antibody based (e.g., Masson trichrome for highlighting connective tissue via collagen hybridizing peptides) (Adv Anat Pathol 2017;24:99)
- Autofluorescence: imaging of natural light emission by tissue slide after light absorption in the absence of fluorophores (Nat Biomed Eng 2019;3:466)
- Coregistration: pixelwise (cellwise) alignment of 2 tissue slides from the same section after tissue restaining (Sci Rep 2014;4:6050)
- Artificial intelligence: computation approaches developed to perform tasks which usually require human intelligence
- Machine learning: computational heuristics that learn patterns from data without requiring explicit programming to make decisions
- Artificial neural networks (ANN): type of machine learning algorithm which represents input data (e.g., images) as nodes; learns image filters (e.g., color, shapes) used to extract histomorphological features; comprised of multiple processing layers that learn representations of objects at multiple levels of abstraction (deep learning); inspired by central nervous system (Nature 2015;521:436)
- Segmentation: use of a computer algorithm for pixelwise assignment of specific classes (e.g., tumor) without specific separation of objects
- Detection: use of computer algorithm to isolate specific objects in an image and report object's bounding box location (e.g., location of nuclei), separating conjoined nuclei, etc. (Am J Pathol 2021;191:1693)
- Generative adversarial networks (GAN): a type of neural network that generates highly realistic synthetic images from input signal (e.g., noise, source image) through iterative optimization of a generator which synthesizes images; this is followed by a discriminator / critique which attempts to distinguish generated from real images
- Paired image translation: requires pixelwise microarchitectural alignment of source stain WSI to target stain WSI; target stain serves as label
- Unpaired image translation: does not require paired tissue stains from same WSI or patient and learns mapping between source and target stains; however, is less precise than paired image translation
- Training cohort: set of cases used for training machine learning model
- Validation cohort: set of cases used for evaluating machine learning model
- Image to image translation: digital conversion of an image from a source domain (e.g., hematoxylin and eosin stained slide) into what it would look like given a target domain (e.g., trichrome stain)
Developing a virtual staining algorithm
- Selection of markers: determined by application (e.g., converting diagnostic H&E stain into trichrome stain for prognostication) and feasibility (i.e., perceived capacity to be inferred from routine stain morphology)
- Primary source staining / imaging modality: WSI of image unstained / stained tissue section, typically using H&E or autofluorescence
- Other source staining / imaging modalities: quantitative phase images, Raman spectroscopy, mIF, IHC
- Common target images: IHC, mIF
- Developing a virtual staining dataset (BME Frontiers 2020;9647163:1):
- Collect specimens with representative tissue architectures (e.g., portal regions, tumor microenvironment, etc.) across a range of diagnostic contexts (e.g., benign versus malignant tissue, different tissue stages, etc.; depends on application); ensure cohorts are well balanced and confounders have been accounted for (e.g., excess adipose tissue, age, sex, etc.)
- Stain and image initial section with source stain (e.g., H&E) across all cases from training cohort
- Stain serial section with target stain (e.g., Ki67) across all cases from training / validation cohort
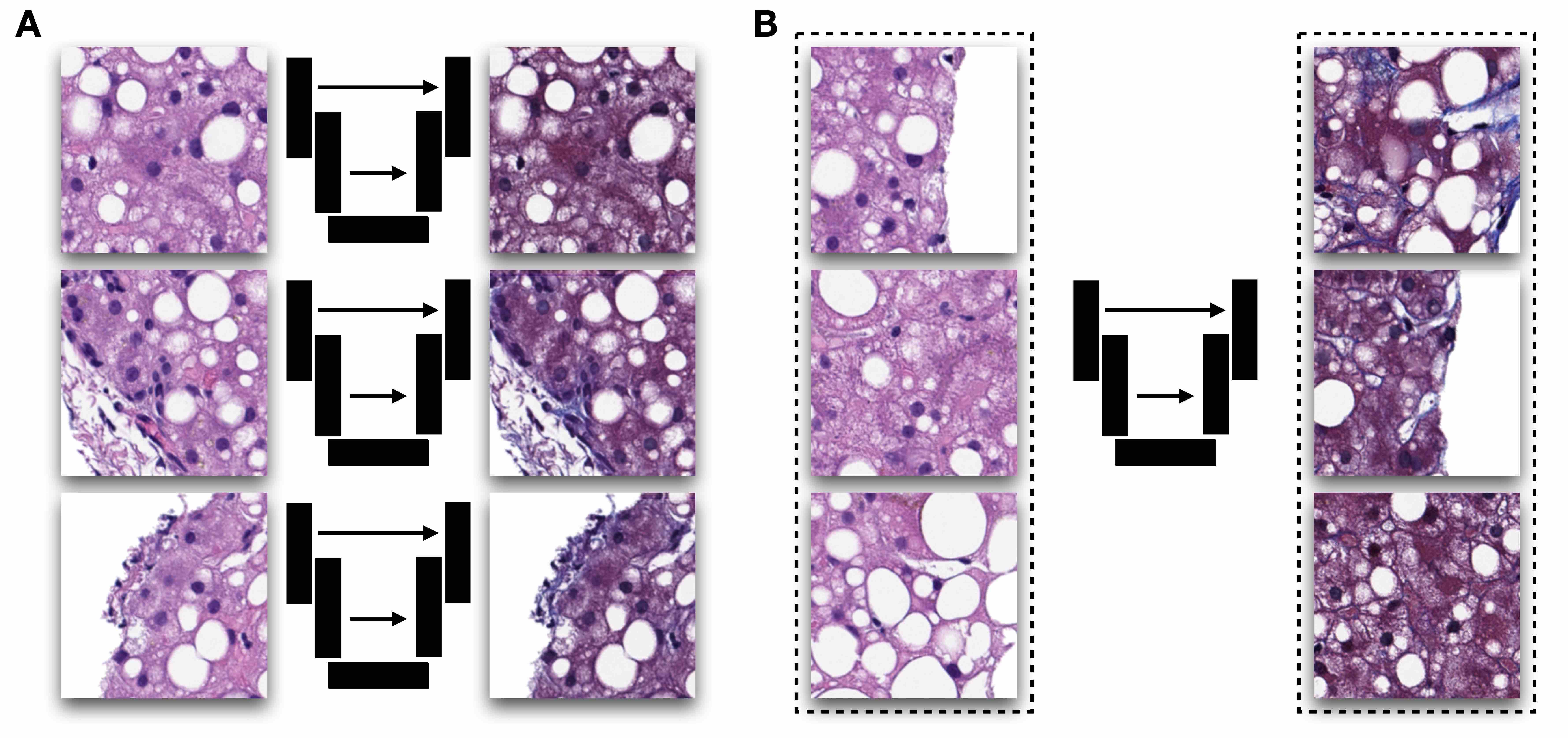
- If performing a paired image translation, source and target stains require application on the same slide (J Pathol Inform 2022;13:100012):
- Most common technique for same slide application is tissue restaining (destaining tissue section, then restaining with second staining application)
- Significant tissue distortion can occur (e.g., nonuniform reductions in tissue size)
- There are approaches which do not require tissue destaining
- Slides must undergo coregistration of nuclear stains (e.g., hematoxylin to SYTO13) or analogous structures (see figures 1, 2)
- If performing an unpaired translation, then there is not a strict requirement for tissue coregistration (i.e., slides stained with target stain do not need to overlap with slides stained with source stain for training the model)
- After segmenting regions of tissue, subimages from source / target WSI are extracted and partitioned into training / validation sets representing source (input) and target (output) stains respectively
- Virtual staining methods (J Pathol Inform 2021;12:43):
- Generative adversarial networks to generate synthetic images of target stain from source using either paired (e.g., Pix2Pix) or unpaired (e.g., CycleGAN) translation methods (see figure 1)
- Pix2Pix generates target stain from source stain by assessing pixelwise concordance in staining between real and synthetic stain and uses discriminator to determine whether synthetic stain was real or fake; requires perfectly aligned images (e.g., pixel to pixel correspondence)
- CycleGAN generates target stain from source by using discriminator to determine whether synthetic stain was real or fake and back translates synthetic stain back to source stain to retain microarchitectural structure via pixelwise concordance; does not require pixelwise registration
- Synthetic images may be postprocessed to yield quantitative measures (e.g., IHC index score)
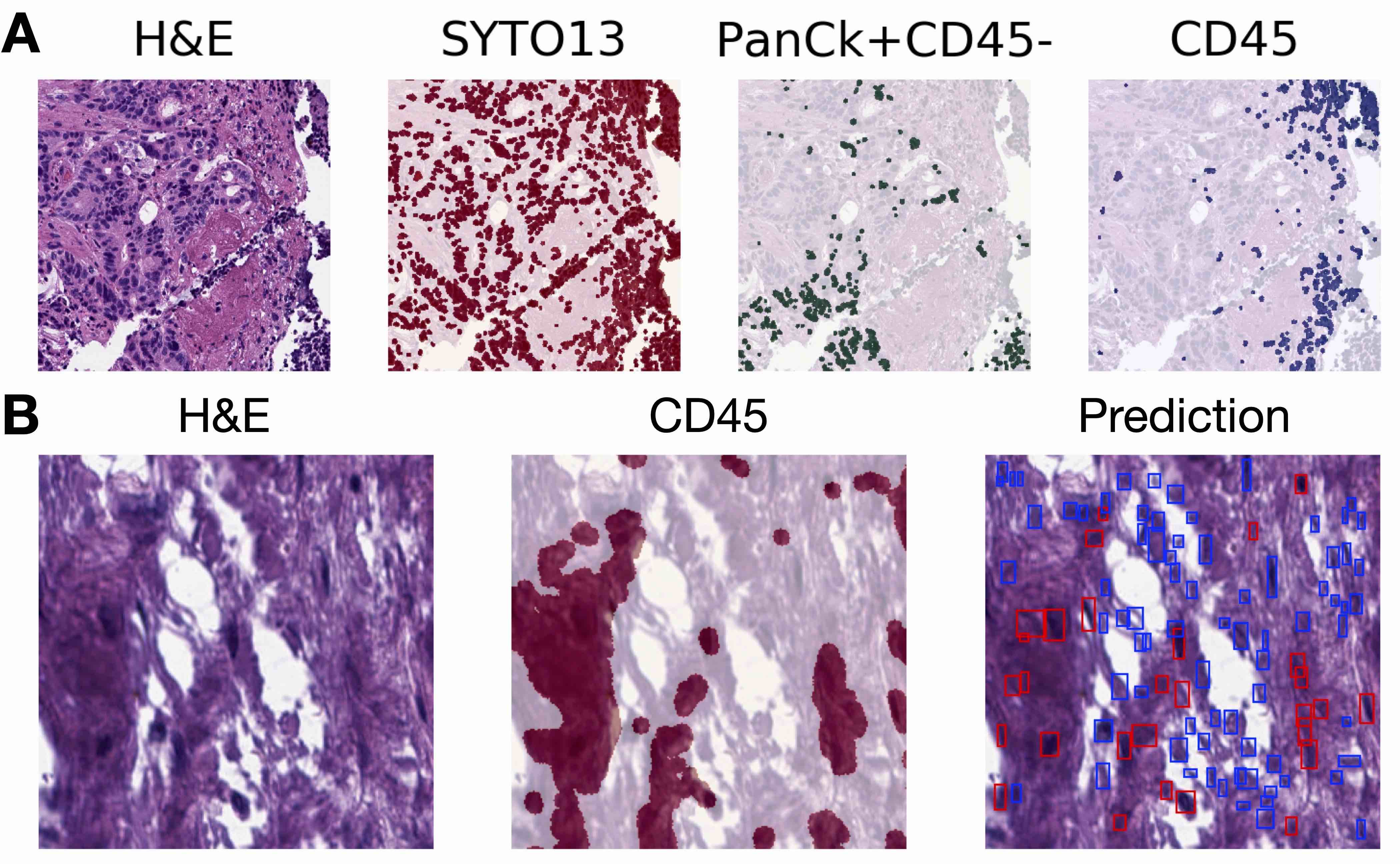
- Segmentation / detection approach:
- Train robust nuclei segmentation / detection algorithms through annotation of nuclei by trained pathologist
- Use stain deconvolution approach (IHC) or intensity thresholding (mIF) to isolate specific target stain / marker (e.g., CD45)
- Overlay marker over nuclei to tag nuclei with label (e.g., labeling immune cells)
- Train separate segmentation / detection algorithm to predict pixelwise presence of specific cell types (e.g., positive / negative staining for SOX10)
- Neural networks are optimized for many iterations as models attempt to converge at optimal mapping between 2 stains
- Generative adversarial networks to generate synthetic images of target stain from source using either paired (e.g., Pix2Pix) or unpaired (e.g., CycleGAN) translation methods (see figure 1)
- Model optimization and selection of optimal virtual stain:
- GAN approach: during training, apply generator to convert select source WSI for collaborating pathologist to visualize intermediate results; training iteration deemed sufficiently concordant to actual staining reagent application is used to convert remaining images
- Segmentation / detection approach: models are optimized based on pixelwise / cell level concordance between predicted and actual stains; best model is selected from training iteration which demonstrates optimal concordance between real / synthetic stain on held out patients
- Additional pathologist annotations (e.g., location of tumor) may help guide virtual staining (e.g., identification of markers colocalized to tumor determined by routine stain)
- Validation, done on a hold out test set that is separate from training / validation set:
- GAN approach: visual assessment for realism and clinical utility (similar ability to grade / stage case) and if paired translation, stain accuracy when postprocessed (i.e., stain separation / accuracy)
- Segmentation / detection approach: pixelwise / cellwise concordance performance (e.g., accuracy) and crosstabulation within macroarchitectures / across the slide
Evaluation of virtual stains
- Visual quality tests (subjective); 2 types of visual quality tests exist:
- Turing test: at random, present pathologists with real and virtual images; Turing test passes if pathologists are unable to distinguish the difference between the real / virtual images
- Staining quality ratings: judging specific tissue architectures (e.g., submucosa) across select slides, pathologists rate quality of tissue staining on a scale from 1 (low quality) to 5 (high quality), noting type and scope of tissue artifacts introduced through virtual staining
- Computational quality test (Brief Bioinform 2022;23:bbac367):
- Inception score (IS) provides an objective measurement of staining quality by using machine learning classifiers on the generated tissue stain to measure its ability to characterize certain tissue architectures (e.g., tumor) while comparing images to ensure a diverse set of images were assessed
- Biomarker quantification (Nat Mach Intell 2022;4:401):
- Marker of accuracy: 1) for paired translation GAN approaches, deconvolving the image into the inferred stains or 2) for segmentation / detection approaches assess agreement with original stains with concordance measurement (e.g., accuracy)
- Quantification of marker across slide: counting cells that stain positive / negative across local region or across slide and correlating with assessment indices (e.g., Ki67 index; area of positive SOX10 staining, etc.)
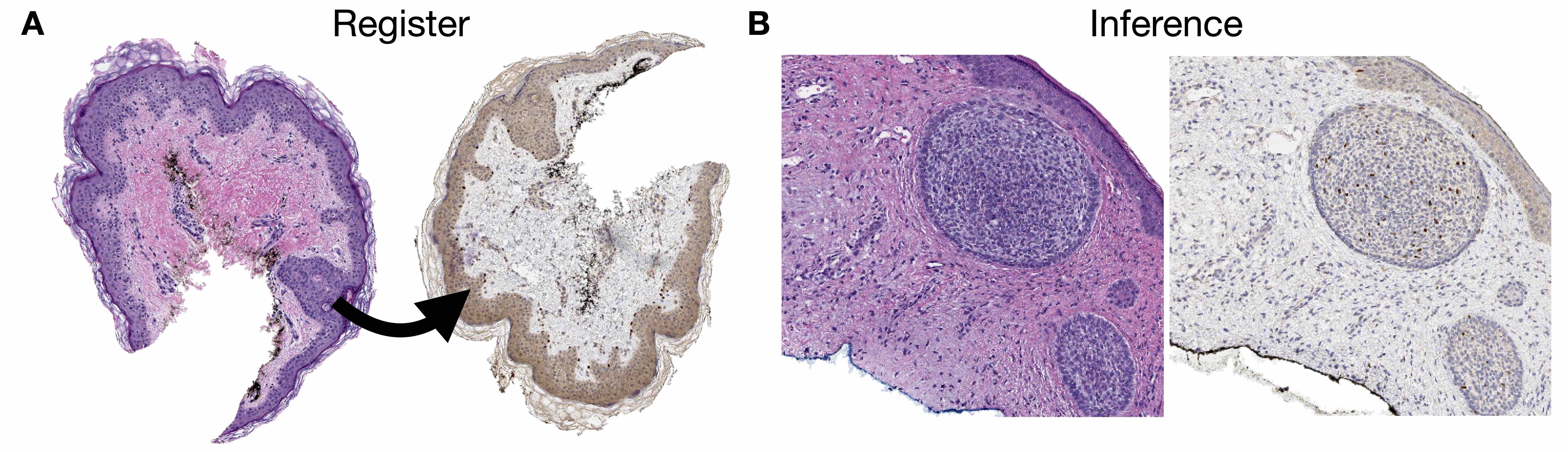
- Clinical concordance for diagnostic / staging practices (Mod Pathol 2021;34:808):
- Noninferiority:
- Can pathologists grade or stage the disease similarly between the real / virtual stain?
- Can be assessed through Kappa agreement, Spearman correlation or regression modeling
- Is the interpretation impacted by artifacts or by perceptions of virtual stain?
- Results should be interpreted in context of how difficult grading / staging can be for this condition (e.g., inter-rater variation) (see figure 3)
- Can pathologists grade or stage the disease similarly between the real / virtual stain?
- Superiority:
- Does virtual staining, when used in conjunction with the standard assessment, permit improved capacity to grade / stage?
- Correlation:
- How well do quantified biomarkers correlate with diagnosis / prognosis?
- WSI from real and virtual stains should be chosen randomly for at least 2 separate assessment periods with a washout period of at least 2 weeks; if real stain is selected for first period, then virtual stain for case / section should be selected for the second period and vice versa; raters should be blinded
- Multiple pathologists should assess the same tissue section with agreement between pathologists and different assessment biases noted
- Noninferiority:
- Correlation with other assays:
- How well does assigned virtual stain stage correlate with other known disease markers (e.g., serology)?
- Is this association more precise than the traditional stain?
- Survival / risk stratification:
- How well does virtual stain biomarker quantification or diagnosis / grade / stage assignments associate with objective markers of recurrence / survival / disease progression (e.g., transplant)?
- Is the association similar or more precise than using the traditional stain?
- Can be assessed through Kaplan-Meier analysis or Cox proportional hazards (i.e., survival) assessment
- Workflow speed and surveys on usage of digital web application to facilitate virtual stain viewing
- Comparative / cost effectiveness comparison with related automation technologies (e.g., autostaining)
Diagrams / tables
Registration software
- Virtual alignment of pathology image series (VALIS)
- OpenCV2
- ViV (Nat Methods 2022;19:515)
- Warpy (Front Comput Sci 2022;3:780026)
- ASHLAR (Bioinformatics 2022;38:4613)
- CODA (bioRxiv 2020 Dec 9 [Preprint])
- HALO
- Visiopharm (Tissuealign)
- ImageJ (TurboReg, UnwarpJ, etc.)
- NiftyReg
- Elastix
- ANTs
- MCMICRO
Virtual staining software
- CycleGAN and Pix2Pix
- Visiopharm (virtual double staining)
- SHIFT
- HistoGeneX
- Owkin
- Indicalabs / HALO
Select example applications of virtual stains
- SOX10 IHC for melanoma (Mod Pathol 2020;33:1638, Front Artif Intell 2022;5:1005086)
- Virtual HER2 IHC staining (Proc SPIE PC12204 ETAI 2022;PC12204:PC122040O, Technical Digest Series 2022;SM5O.2)
- Label free staining from tissue autofluorescence into H&E, Jones and trichrome (Light Sci Appl 2020;9:78, Light Sci Appl 2019;8:23, Nat Biomed Eng 2019;3:466)
- Gleason scoring (Sci Rep 2022;12:9329)
- Pancytokeratin, tumor stromal ratio and p53 (Sci Rep 2020;10:17507, Sci Rep 2021;11:192557, NPJ Precis Oncol 2022;6:14)
- Special stains (e.g., periodic acid-Schiff) (Nat Commun 2021;12:4884)
- Ki67 (IEEE Trans Med Imaging 2021;40:1977)
- PAS stain (kidney) (J Pathol Inform 2022;13:100107)
- IHC to mIF (e.g., DAPI, Lap2, BCL2, BCL6, CD10, CD3 / CD8, Ki67) (Nat Mach Intell 2022;4:401)
- Tumor microenvironment characterization (Proc SPIE 12039 Medical Imaging 2022;12039:120391H)
- Adipocytes (nuclei, lipid, cytoplasm) (PLoS One 2021;16:e0258546)
- Intraoperative virtual staining (Nat Biomed Eng 2022 Sep 19 [Epub ahead of print])
- Virtual staining of skin biopsies from reflectance confocal microscopy (Light Sci Appl 2021;10:233)
- Large scale clinical validation of virtual trichrome staining (Mod Pathol 2021;34:808)
- Virtual staining of distinct cellular components (e.g., endosome, actin, golgi, etc.) (Sci Adv 2021;7:eabe0431)
- Fibrosis quantification (Med Image Anal 2022;81:102537)
- Blood smears (BME Frontiers 2022;9853606:1)
- Mitosis detection via PHH3 stain (2020 IEEE 17th ISBI 2020;1770)
- Other examples (BME Frontiers 2020;9647163:1)
Advantages and disadvantages
- Advantages (Nat Biomed Eng 2019;3:466):
- Ameliorate laboratory infrastructure burdens through quick access to reflexive tissue staining at virtually no cost
- Improves precision and tedious quantification of biomarkers
- Can provide access to stains with reasonable multiplexing to facilitate spatial molecular assessment
- Can make remote pathology consultation more practical due to reliance on digital assessment
- Potentially can be employed to standardize the application of staining reagents
- Virtually stained tissue can be used to augment datasets for training downstream machine learning workflows (e.g., IHC marker quantification) (see figure 4)
- Disadvantages:
- GANs may hallucinate histological features that look real but are incorrectly placed or should not be present; this often occurs when sections are imprecisely aligned for paired translation techniques and sometimes when using unpaired translation techniques
- Without evaluating virtual stains on generated images of WSI (e.g., looking only at representative subimages), it is possible to choose a virtual staining model that over / under stains
- Tissue artifacts and variable slide staining can introduce aberrant staining in virtual slides; multicenter validation or finetuning of technology is recommended for demonstrating broad application
- Depending on the application, disagreement between raters may make assessment challenging, reducing the study power by adding noise
- Pixelwise agreement between real / virtual stain does not necessarily reflect the impact on real world grading / staging
- Turing test can be easily fooled by presenting pathologists with smaller image crops (i.e., tissue resolution, larger images have higher likelihood of containing an artifact) or not enough images to draw inferences (i.e., sample size); does not accurately reflect impact on real world grading / staging
Board review style question #1
As opposed to an unpaired image translation task, what different set of procedures are absolutely required to obtain data for a paired image translation task?
- Application of at least 2 different tissue stains to the same tissue section
- Segmentation of tissue and partitioning into training / validation sets
- Tissue restaining and image coregistration of same tissue section
- Tissue staining of a partially overlapping set of patients
Board review style answer #1
C. Tissue restaining and image coregistration of same tissue section. C is the only true statement. While any of these steps are helpful for data acquisition for paired and unpaired tasks, a paired task can only be performed if there is precise microarchitectural alignment to the same tissue section. While A is also true, it does not explicitly discuss the coregistration procedure. Procedures discussed in D are relevant for an unpaired analysis. Partitioning the dataset, as discussed in B, can be done for both unpaired and paired assessments. See image above for an example of processing required for a paired analysis.
Comment Here
Reference: Virtual staining
Comment Here
Reference: Virtual staining
Board review style question #2
Which assessment method for virtual staining technologies is aligned with clinical application / validity of these technologies?
- Concordance with stage / serology / survival
- Cost effectiveness comparison
- Surveys for attitudes and beliefs surrounding adoptability
- Virtual assessment time
Board review style answer #2
A. Concordance with stage / serology / survival. While all of these assessment methods are relevant for determining the potential to adopt a virtual staining technology, A demonstrates real world application of this approach. If assessments of virtually stained slides reduces the capacity to grade / stage, the approach should not be adopted or should be further iterated on until it can demonstrate noninferiority to the current assessment method. Other visual assessment methods include the Turing test and inception score, which can help determine visual fidelity. The other mentioned assessment methods are helpful for determining the potential to adopt the technology (i.e., does this technology reduce costs [B] and the time required to stain / assess [D]); surveys discussed in C are opportunities to solicit feedback on changes that can make the technology more user friendly and identify additional barriers for adoption. While virtual staining technologies are used to generate synthetic images of tissue, application of these assessment methods are helpful for demonstrating the utility of these technologies. These assessments should be performed in large scale cohort settings prior to initiating a multicenter randomized controlled trial.
Comment Here
Reference: Virtual staining
Comment Here
Reference: Virtual staining