Table of Contents
Definition / general | Essential features | Terminology | Common cellular morphometric features for cytological assessment | Developing a cytological assessment algorithm | Evaluation of computer algorithms | Select examples of cytopathology image analysis approaches | Model training tips | Advantages & disadvantages | Software for automated cytopathology image analysis workflows (Python) | Diagrams / tables | Videos | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Levy J, Vaickus L. Automated assessment of cytology specimens. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/informaticsautoassesscytology.html. Accessed May 13th, 2024.
Definition / general
- Automated assessments of cytology specimens use computer algorithms to quickly and quantitatively assess cells in a cytology slide
- Assessments provide an overall estimate of atypia or locate abnormal cells
- Image recognition software, especially artificial neural networks (ANN), are often used for these assessments
- Automated assessments can reduce burden (e.g., fatigue, variability, misdiagnoses, costs) in cytology screening practices, especially in areas that use outdated methods and lack subspecialists
Essential features
- Automated cytology assessments use computer vision software and artificial neural networks to quickly evaluate thousands of cells in a cytology specimen for the presence and extent of abnormal cells that may indicate atypia (J Am Soc Cytopathol 2019;8:230, Biomed Eng Comput Biol 2016;7:1)
- These assessments can improve the efficiency of cytopathology by allowing for the rapid evaluation of many cytology slides, which are traditionally time consuming and prone to error (Acta Cytol 2021;65:301)
- Accuracy of the results may be affected by factors such as the quality of the specimen, the type of specimen, the method of collection, the staining and fixation and other factors such as infections, hematuria and stones (Acta Cytol 2021;65:301)
- Techniques for automated assessment include cytometry, image recognition software and machine learning algorithms (Med Image Anal 2023;84:102691)
- Automated assessments are not meant to replace traditional cytopathology and should be reviewed by cytologists for confirmation of findings
Terminology
- Whole slide image (WSI): digitized representation of tissue slide after whole slide scanning at 20x or 40x resolution (e.g., using Aperio AT2 or GT450 scanner); slide dimensionality can exceed 100,000 pixels in any given spatial dimension
- Subimage / patch: smaller, local rectangular region extracted from a WSI, often done to reduce the computational resources required for the development and deployment of machine learning algorithms (J Pathol Inform 2019;10:9)
- Z stacking: technique that combines multiple images / focus at different depths to create a 3D representation of the cells in a sample
- SurePath / ThinPrep: 2 methods for collecting and preparing cytology samples (Cytopathology 2021;32:654)
- ThinPrep imaging system: software approved by the Food and Drug Administration (FDA) for screening Pap smear slides for cervical cancer by identifying abnormal cells (Diagn Cytopathol 2021;49:559)
- Nuclear to cytoplasm ratio (N:C ratio): ratio of area of nucleus as compared to cytoplasm in cell (Cancer Cytopathol 2015;123:524)
- Higher N:C ratio suggests cellular abnormality though is dependent on cell type
- Hyperchromasia: increased pigment in nucleus of cell, causing nucleus to appear darker under microscope, suggestive of abnormality (Acta Cytol 2020;64:511)
- Frayed chromatin: refers to type of chromatin pattern in cell where chromatin is unraveled, suggestive of abnormality (Diagn Cytopathol 1986;2:76)
- Nuclear grooves: indentations in nucleus of a cell that may suggest DNA damage
- Squamous cell differentiation: refers to the degree in which a squamous cell resembles a basaloid structure; particularly important for evaluation of Pap smear specimens (Comput Methods Programs Biomed 2013;111:128)
- Cell clusters: group of cells in close arrangement (Cancer Cytopathol 2023;131:19)
- Follicular cells tend to aggregate for thyroid cytology specimens and are the primary object of interest
- Whereas in urine specimens, clusters are more likely to result from specimen type and preparation method, though fibrovascular structures and atypical clusters may factor into overall indicators of malignancy
- The Paris System (TPS): semisubjective quantitative assessment system for evaluating high grade urothelial carcinoma (J Pers Med 2022;12:170)
- Bethesda system: standardized system for interpreting cytology specimens, particularly for reporting on specimen atypia for thyroid, salivary glands and cervix cytopathology (Acta Cytol 2017;61:359)
- Fine needle aspiration: use of thin needle to collect samples of cells from tissue / organs
- Brush: use of small brush to collect cells from tissue surface
- Voided specimen: urine sample collected by urinating into a container; exfoliative
- Connected components: technique for separating individual cells or objects within an image after intensity thresholding to remove background whitespace; based on contiguous pixel elements that exceed a minimum size threshold
- Artificial intelligence: computation approaches developed to perform tasks which typically require human intelligence
- Machine learning: computational heuristics that learn patterns from data without requiring explicit programming to make decisions
- Artificial neural networks (ANN): type of machine learning algorithm that represents input data (e.g., images) as nodes
- Learns image filters (e.g., color, shapes) used to extract histomorphological / cytological features
- Comprised of multiple processing layers to represent object at multiple levels of abstraction (deep learning)
- Inspired by central nervous system (Nature 2015;521:436)
- Classification: use of computer algorithm for assigning type of object; useful for sorting out different objects in cytology specimen (e.g., debris) (Cancer Commun (Lond) 2020;40:154)
- Clustering: use of computer algorithm to group objects together, either based on similar features or spatially based on their colocalization
- Segmentation: use of a computer algorithm for pixelwise assignment of specific classes (e.g., nucleus) without specific separation of objects
- Detection: use of computer algorithm to isolate specific objects in an image and reporting of object's bounding box location (e.g., location of nuclei, cytoplasmic borders), separates conjoined nuclei, etc. (Am J Pathol 2021;191:1693)
- Generative adversarial networks (GAN): type of neural network that generates highly realistic synthetic images from input signal (e.g., noise, source image) through iterative optimization of generator, which synthesizes images and discriminator / critique which attempts to distinguish generated from real images (Mod Pathol 2021;34:808)
- Evaluation metrics: used to depict performance of automated cytology algorithm
- Accuracy: proportion of observations that were classified correctly
- Sensitivity: proportion of cases that were classified correctly at the given cutoff probability threshold
- Specificity: proportion of controls which were classified correctly at the given cutoff probability threshold
- AUC: area under the receiver operating characteristic curve, an overall measure of performance considering sensitivity / specificity reported across many cutoff thresholds
- F1 score: captures tradeoff between sensitivity / specificity
- Intersection over Union (IoU): used to evaluate accuracy of cell localization algorithm by comparing the area overlap between the predicted cell location and ground truth location to the area union between the predicted / ground truth location
- Mean average precision (mAP) averages the precision of the model across many IoU thresholds (i.e., minimum IoU between predicted / ground truth locations to indicate a true positive detection)
- Training cohort: set of cases used for training machine learning model
- Validation / test cohort: set of cases used for evaluating machine learning model
Common cellular morphometric features for cytological assessment
- Types of pertinent algorithms / metrics used for these assessments are denoted in the subbullet points (IEEE J Biomed Health Inform 2014;18:94, Cancer 1998;84:115, J Cytol 2011;28:98)
- Cell size: as defined by the combined cytoplasmic and nuclear area; abnormal cells may not conform to the typical cell type size
- Connected components analysis and cell detection algorithms are first used to separate cells
- Area and perimeter are common size measurements
- Cell shape: abnormal cells tend to have irregular / abnormal shapes
- Eccentricity, inferred from the major and minor axis lengths
- Other measurements that may be used to characterize cell shape include circularity, elongation, solidity and compactness, among others
- Nucleus area: see N:C ratio
- N:C ratio can be calculated using segmentation neural network or GAN, both of which can predict pixelwise presence of nucleus and cytoplasm
- Nucleus shape: see cell shape; abnormal nuclear shapes may indicate specimen atypia
- Abnormal nuclear / chromatin features: see frayed chromatin or chromatin coarseness
- Convolution or image filters may be used to identify abnormal nuclear or chromatin features
- Convolutional neural networks (CNNs) may also be used to identify and classify abnormal nuclear or chromatin features; potentially labeled by cytopathologist as present / absent or on some measurement scale
- Cytoplasmic features: examples of abnormal cytoplasmic features include presence of vacuoles / inclusions and granules where not expected
- Measures that may be used to characterize cytoplasmic features include intensity, color and texture in addition to neural network methods
- Cell / object type: useful for screening out cell types typically irrelevant for specific cytological assessment (e.g., bladder cancer - identify urothelial cells, cervical cancer - identify squamous cells, thyroid cancer - identify follicular cells, etc.)
- Machine learning algorithms or other computational techniques may be used to classify cells based on their type
- Dense / abnormal architecture: useful for identifying clusters of cells with dense overlapping cytoplasmic borders which may signify specimen atypia
- Image analysis algorithms (e.g., detection models) may be used to identify dense or abnormal cell clusters
- Subjective measure of atypia: combination of features leading to a cytopathologist determination of cytological atypia (Cancer Cytopathol 2019;127:98)
- Cytopathologists may use a combination of morphometric features, such as cell size, shape and chromatin characteristics, to subjectively evaluate the atypia of a cytology specimen
- Cell size: as defined by the combined cytoplasmic and nuclear area; abnormal cells may not conform to the typical cell type size
Developing a cytological assessment algorithm
- Cohort selection: perform a retrospective review of cases in the institution
- Ensure diagnoses are up to date using latest cytological screening criteria; this may require reexamination from multiple cytopathologists
- Decide on patient outcome measures, usually degree of specimen atypia / diagnosis for cytology screening
- Recurrence and death as endpoints have become increasingly relevant though requires significant follow up time
- Select cases and controls based on study design type (Nephron Clin Pract 2009;113:c214, Plast Reconstr Surg 2010;126:2234)
- Retrospective cohort design: all or random subset of controls and cases are selected across a well defined study period
- Case control study: select individuals based on outcome status
- Allows for case control matching, optimal for rare events
- Cannot report negative / positive predictive value as this requires knowledge of disease prevalence
- Record relevant patient characteristics (e.g., review date, specimen type, specimen preparation, presence of slide artifact, age, sex, histological follow up, history of chemotherapy, hematuria, diagnosis, etc.)
- Divide cases and controls into training and validation cohorts
- Ensure any repeat exams / measures for specific patients are included in the same cohort to avoid endogeneity (i.e., linking information from the training to the validation / test set, reducing the capacity to generalize the study findings)
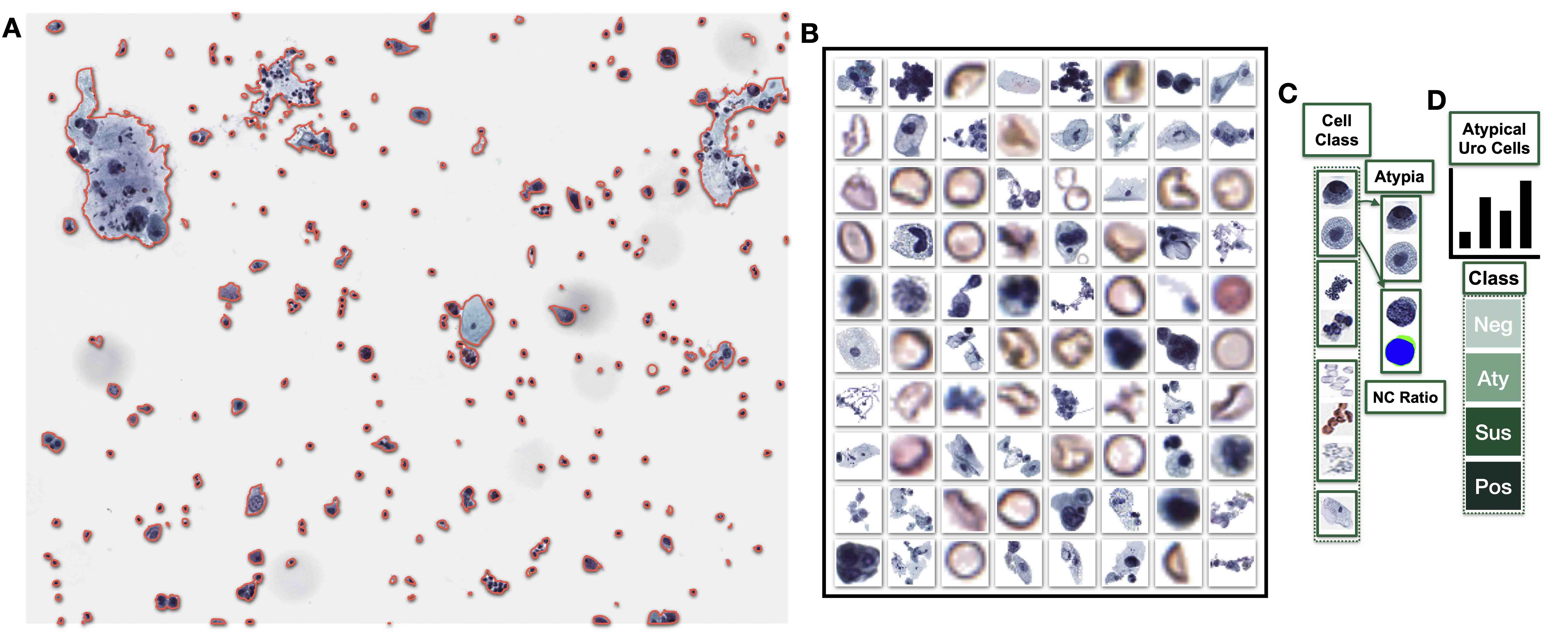
- Developing cell and cluster level algorithms (see Figure 1)
- Use connected components analysis or detection models to isolate cells and cell clusters within training cohort; randomly partition to training and validation cohorts
- Cells belonging to the same patient should be partitioned into the same cohort
- Used to calibrate cell / cluster level models / scores for external application on the test set
- Isolate individual cells from cell clusters if appropriate; this may require generating a training and validation set of clusters trained on pathologist annotations localizing cell types within adjacent / bordering cells
- Validated based on 1) IoU and mAP statistics, which assess overlap between predicted and actual cell locations; 2) cluster level metrics (e.g., correlation to ground truth based on number of cells / abnormal cells identified within each cluster)
- Train and validate several machine learning models or computer vision methods to derive cell level scores
- Requires cytopathologists to place cell types into categories if classifying / filtering cell types and outline nucleus / cytoplasm if calculating the N:C ratio
- Cell level scores can be aggregated to the cluster level to form cluster level scores
- E.g., number of abnormal cells, area of dense architecture
- E.g., essential for assessment of follicular carcinoma
- Use connected components analysis or detection models to isolate cells and cell clusters within training cohort; randomly partition to training and validation cohorts
- Form slide level scores based on aggregate cell and cluster predictors; calculate for each slide across training / validation cohorts
- May require developing thresholds for what constitutes a benign / malignant cell, etc. and either sum or calculate the proportion of cells exhibiting a specific property
- E.g., number of urothelial cells that are atypical
- Train and validate machine learning model on slide level scores to predict specimen atypia
- Incorporate patient characteristics (e.g., age, sex and medical history)
- Validate using AUC: measures the model's ability to distinguish between true positive and true negative cases
- Ensure slides within patient are partitioned to the same training / validation / test set - avoids endogeneity (i.e., linking information from the training set to the validation / test set, which can reduce the ability to generalize the study findings)
- Assess for importance of specific predictors through model interpretation
- Importance of specific predictors (e.g., cell size, shape or nuclear features) in predicting specimen atypia can be determined through feature importance, partial dependency plots or regression coefficients
- Develop web application and helpful displays to facilitate rapid and complete examination
- Highlight areas of atypia or provide other relevant information to the user
- Package software for distribution
- Making software more reproducible (PLoS Comput Biol 2017;13:e1005412, Nat Biotechnol 2017;35:342, Nat Biotechnol 2017;35:316, PLoS Comput Biol 2022;18:e1009823)
Evaluation of computer algorithms
- Cell level algorithm: F1 score, sensitivity, specificity, AUC are all helpful metrics for determining whether cells and their atypia are appropriately classified (J Intern Med 2020;288:62)
- Cluster level algorithm: IoU / mAP reported within / across slides; correlate cluster level measures using correlation statistics or AUC
- Slide level algorithm: validate using the concordance statistic (e.g., AUC), comment on sensitivity / specificity for cutoff threshold; negative and positive predictive values cannot be reported for case control study designs
- Association with clinical endpoints: establish association of predicted slide level scores with specimen atypia using either hierarchical logistic or ordinal regression models; correlate each slide level feature (e.g., number of atypical cells) with these endpoints in addition to other relevant endpoints (e.g., molecular findings, etc.)
- Survival / risk stratification: how well does the overall specimen atypia estimate associate with objective markers of recurrence / survival / disease progression (e.g., transplant); is the association similar or more precise than using traditional cytological examination
- Can be assessed through Kaplan-Meier analysis or Cox proportional hazards (i.e., survival) assessment (World J Urol 2016;34:1405)
- Surveys on usability of the tool and feedback on visual displays (NPJ Digit Med 2020;3:107)
Select examples of cytopathology image analysis approaches
- Initial applications of computers for cellular analysis (Trans N Y Acad Sci 1955;17:250, Experientia 1955;11:163, J Pathol 2019;249:286, Best Pract Res Clin Obstet Gynaecol 2011;25:585)
- Advanced imaging techniques (Diagn Cytopathol 2019;47:5)
- Challenges developing AI systems for cytological assessment (Acta Cytol 2021;65:301)
- Reviews of digital cytology methods (Acta Cytol 2011;55:227, Med Image Anal 2023;84:102691, Cytojournal 2008;5:16, J Am Soc Cytopathol 2019;8:230, Biomed Eng Comput Biol 2016;7:1, Monogr Clin Cytol 2020;25:91)
- Whole slide imaging (Cancer Cytopathol 2017;125:701, Cytopathology 2020;31:372)
- Urine cytopathology (see Figure 1)
- AI algorithm for urine cytology (Cancer Cytopathol 2019;127:658, J Urol 1998;159:1619, Br J Urol 1998;81:574, Cytometry A 2021;99:732, BJU Int 2022;130:235, Cancer Cytopathol 2021;129:984, Cancer Cytopathol 2019;127:218)
- Examples of dataset curation (Acta Cytol 2022;66:46)
- Alignment with histological findings (Front Oncol 2022;12:901586)
- Evaluation of algorithms (Cancer Cytopathol 2022;130:872)
- Development of atypical burden score (Cancer Cytopathol 2019;127:98)
- Processing of cell clusters (Cancer Cytopathol 2023;131:19)
- Hyperchromatic nuclei (Cancer Cytopathol 2022;130:363)
- Generating synthetic images of urothelial cells (J Am Soc Cytopathol 2022;11:123, J Am Soc Cytopathol 2023;12:126)
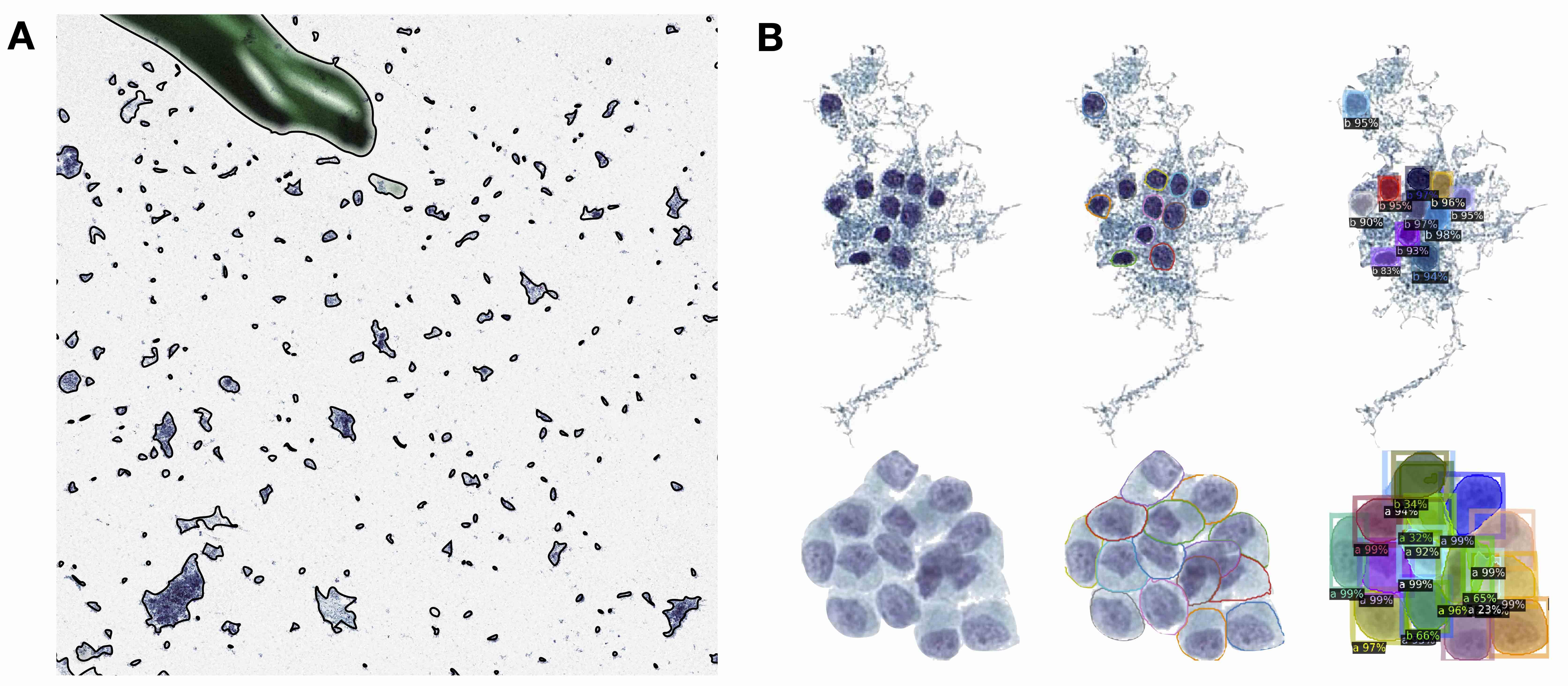
- Thyroid cytopathology (see Figure 2)
- Machine learning applications (Arch Pathol Lab Med 2022;146:872, J Pathol Inform 2022;13:100004, J Cancer 2019;10:4876, Cancer Cytopathol 2020;128:287, Med Image Anal 2021;67:101814, J Pathol Inform 2018;9:43, Cancers (Basel) 2021;13:3891)
- Companion ancillary tool to molecular testing for indeterminate atypical cases (Cancer Cytopathol 2023;131:217)
- Cytological features (J Pathol Inform 2019;10:29)
- Review (Cytopathology 2020;31:432)
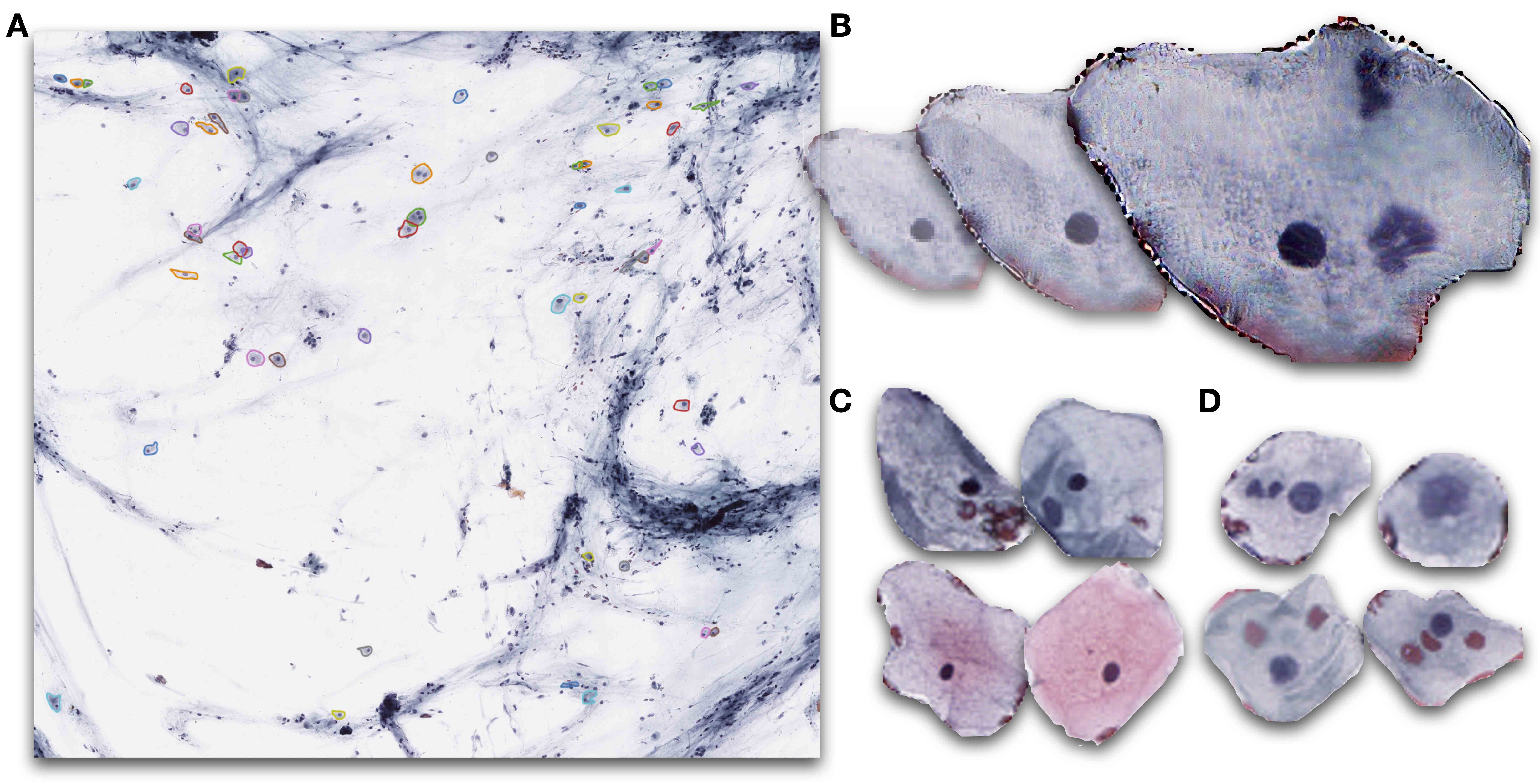
- Cervical cytopathology (see Figure 3)
- Lessons from implementing Pap test screening algorithm (Acta Cytol 2021;65:286)
- Manual versus automation assisted reading (Lancet Oncol 2011;12:56, Gynecol Oncol 2007;104:134, Diagn Cytopathol 1999;21:296, J Reprod Med 1999;44:11, Acta Cytol 2000;44:128)
- HPV screening with multiple stains (J Natl Cancer Inst 2021;113:72)
- Large scale observational / trial study with AI automation (J Natl Cancer Inst 2019;111:923, Gynecol Oncol 2020;159:171, Cancer Med 2020;9:6896, Acta Cytol 2020;64:486)
- Deep learning approaches integrating HPV information with cytology (Cancers (Basel) 2022;14:1159, Int J Med Inform 2022;159:104675, Nat Commun 2021;12:5639)
- Global health applications (JAMA Netw Open 2021;4:e211740)
- Review of automated Pap smear screening (Diagn Cytopathol 2019;47:20, NPJ Digit Med 2022;5:19, Front Oncol 2022;12:851367, Cancer Cytopathol 2022;130:402)
- Commercial systems (J Pathol Inform 2022;13:100095, Acta Cytol 2014;58:469)
Model training tips
- Stain normalization: standardization of staining reagents is essential for ensuring reproducibility of study findings and can be accomplished using Macenko or generative adversarial network (GAN) based methods, among others (Comput Struct Biotechnol J 2021;19:3852, Front Med (Lausanne) 2021;8:746307, Cytometry A 2022;101:1068)
- Conversion between stains for external application of stains for collaborating research institutions (Heliyon 2021;7:e06331)
- Augment dataset: provides additional, unique training samples which can improve model performance
- Data augmentation techniques: includes randomly changing stain properties, rotating / distorting images, etc. to resemble unseen cytology images (Med Image Anal 2019;58:101544)
- Neural networks that produce synthetic cytology images to increase the number of training samples (Proc Mach Learn Res 2019;116:10)
- For teaching and quality assurance (J Am Soc Cytopathol 2022;11:123, J Am Soc Cytopathol 2023;12:126)
- Transfer learning: developing a neural network on a set of related cytopathology images (e.g., thyroid), then continue to train or finetune on the specific set of related cytological images (e.g., urine) (Diagnostics (Basel) 2022;12:2477, IEEE J Biomed Health Inform 2017;21:1633)
- Methods to rapidly annotate cytology images
- Bayesian active learning: iteratively labeling images based on algorithmic uncertainty / mistakes (Sci Rep 2019;9:14347)
- Unsupervised clustering: sorts cells into groups and suggests groups to annotate (J Pathol Inform 2022;13:100146, Nat Commun 2012;3:1032, Am J Clin Pathol 2022;157:5)
- Point annotations: avoids need to outline objects in WSI (Med Image Anal 2020;65:101771, IEEE J Biomed Health Inform 2021;25:1673, IEEE Trans Med Imaging 2020;39:3655)
- Super resolution (IEEE Trans Med Imaging 2020;39:2920, IEEE Trans Med Imaging 2022;41:383 Appl Soft Comput 2022;115:108208)
- Resolve disagreements between cytopathologists or incorporate multiple ratings during model evaluation
- Consultation with biostatistician to set up multilevel modeling evaluation framework
- Interpretation techniques, such as SHAP and GradCAM, highlight important cell features and can be used to determine systematic sources of error (Nat Mach Intell 2020;2:56)
Advantages & disadvantages
- Advantages of automated cytology assessment (Nat Rev Clin Oncol 2019;16:703, Vet Clin North Am Small Anim Pract 2023;53:73)
- Efficiency: rapid and complete analysis of large numbers of specimens
- Accuracy: less prone to error compared to manual assessment
- Objectivity: algorithms developed based on objective diagnostic criteria, reducing subjectivity and interobserver variability
- Workload: reduces workload for cytopathologists, allowing them to focus on more complex / challenging cases
- Can improve access to care in low resourced regions by either freeing up subspecialist time or reducing barriers to entry for nonspecialists
- Disadvantages of automated cytology assessment (Acta Cytol 2018;62:68, Acta Cytol 2021;65:301)
- Dependence on imaging technologies: may be prone to failure / malfunction that would require opting for the manual assessment
- May be impacted / less accurate with poor quality or abnormal specimens (e.g., deficiencies in specimen preparation or the presence of blood, imaging artifact, pixelation of cells)
- May not account for Z stacking or 3D cell modeling
- Training: likely requires time consuming and resource intensive training to familiarize use of the technology
- May not be preferred by cytopathologists unfamiliar with digital technologies as compared to analog methods, potentially leading to misuse of the technology
- Requires significant computing infrastructure investment
- False negatives: may not identify all abnormalities, potentially leading to missed diagnoses
- External application: may not be applicable to other institutions with different patient characteristics and specimen preparation methods
- Dependence on imaging technologies: may be prone to failure / malfunction that would require opting for the manual assessment
Software for automated cytopathology image analysis workflows (Python)
- ASAP, Qupath, ImageJ: used for annotating / isolating cytology images (Sci Rep 2017;7:16878, Comput Struct Biotechnol J 2021;19:852)
- Scikit-image, OpenCV2: image analysis frameworks (PeerJ 2014;2:e453, Comput Biol Med 2017;84:189)
- Scikit-learn: machine learning framework (J Pathol Inform 2022;13:100004)
- PyTorch, Keras, Tensorflow: deep learning frameworks (Mol Cancer Res 2022;20:202)
- Detectron2, MMDetection: cell detection frameworks (Cancer Cytopathol 2023;131:19)
Diagrams / tables
Videos
AutoParis-X Demo Tutorial
Board review style question #1
What is the primary purpose of using connected component / detection image analysis to isolate cells and cell clusters?
- To identify the location of cells within whole slide image
- To identify the number of abnormal cells within a cluster
- To identify the number of cells within a cluster
- To identify the type of cells within cluster
Board review style answer #1
A. To identify the location of cells within whole slide image (WSI). While answers A - D require isolation of cells and cell clusters, the primary purpose is to identify the locations of cells / clusters within WSI for further analysis. Answers B, C and D are specific to cell clusters but do not discuss the isolation of individual cells within the specimen, only orthogonal tasks (e.g., counting cells after they have been isolated). Furthermore, convolutional neural networks and segmentation neural networks are effective at predicting the type and subjective / objective atypia of cells but are not as capable at localizing cells as compared to detection approaches. Before any analysis can be done, cells must be isolated using connected components / detection analyses (A). See Figure 1A for an example of cell / cluster localization prior to downstream analysis.
Comment Here
Reference: Automated assessment of cytology specimens
Comment Here
Reference: Automated assessment of cytology specimens
Board review style question #2
What is the primary function of the intersection over union (IoU) and mean average precision (mAP) statistics in the context of cell localization?
- To assess the overlap between predicted and actual cell locations
- To calculate the nuclear to cytoplasm (N:C) area ratio
- To determine the degree of squamous cell differentiation
- To measure the correlation between predicted and actual cluster level metrics (e.g., number of abnormal urothelial cells)
Board review style answer #2
A. To assess overlap between predicted and actual cell locations. IoU and mAP are both detection model performance measures. The goal of a detection model is to localize cells given an input image containing multiple cells. The nuclear to cytoplasm ratio (B) can be calculated using a segmentation neural network (key evaluation statistics include the F1 score and AUC). Cluster level metrics (D) can be correlated using the Spearman correlation coefficient and is not related to cell localization but cluster level metrics can be derived from detected cells. Squamous cell differentiation (C) is unrelated to these localization statistics.
Comment Here
Reference: Automated assessment of cytology specimens
Comment Here
Reference: Automated assessment of cytology specimens